Proceedings of The World Avocado Congress III, 1995 100 - 105
PROGRESS OF THE STUDY ON THE GENETIC RESOURCES OF AVOCADO IV. USE OF
THE SPACER SIZE VARIATION OF RIBOSOMAL 5S GENES TO IDENTIFY THE HORTICULTURAL
RACE OF AVOCADO GENOTYPES
J. Fiedler and G. Bufler
Institut für
Obst-, Gemüse- und Weinbau (370)
Universität
Hohenheim
70593 Stuttgart
Germany
Abstract
'Universal' primers
have been used to amplify the nontranscribed spacer (NTS) of the ribosomal 5S
gene of avocado genotypes by the polymerase chain reaction (PCR). A rich size
heterogeneity of NTSs of individual avocado genotypes was detected after high
resolution polyacrylamide gel electrophoresis and silver staining. Specific
sizes of NTS seem to be common for members of each horticultural race of
avocado and may be used as a diagnostic tool to identify the horticultural race
of avocado genotypes.
1. Introduction
Three horticultural or ecological races of avocado (Persea americana
Mill.) can e distinguished, namely Mexican, Guatemalan, and West Indian;
Bergh and Ellstrand (1987) gave a detailed typification of these groups. It is
assumed, that the majority of avocado cvs. are 'pure' to each race, but that
some of them may be actually hybrids of yet unknown lineage (Lahav and Gazit,
1994).
It is widely
acknowledged that the heterogeneity of 5S ribosomal DNA (rDNA) an provide a
useful tool in plant systematic studies (Sastri et al., 1992). In higher plants
the genes that code for 5S rRNA are organized into clusters of tandem repeats.
Every repeat unit consists of a transcribed region of approximately 120 base
air (bp) and a NTS region varying in size between 100 bp and 700 bp (Sastri et
al., 1992). It is this variation in size (and sequence) which makes 5S rDNA so
suitable or plant systematic studies. The purpose of this study was to test the
variation of NTS sizes between genotype
representing the three horticultural races of avocado, and to determine whether
it could be used as a diagnostic tool to identify the horticultural race of an
avocado genotype. NTSs of avocado cvs. were selectively amplified using the PCR
and 'universal' primers as applied previously to 5S genes of barley (Kolchinsky
et al., 1991) and wheat (Cox et al., 1992).
2. Material and methods
2. 1. Plant material
Avocado leaf samples of cultivars (cvs.) or germplasm bank accessions
were collected in Israel (The VoIcani Center, Bet-Dagan) or in Mexico
(CICTAMEX, Coatepec Harinas, Mexico), lyophilized or oven-dried and sent to the
University of Hohenheim for analysis.
2.2. DNA
extraction
DNA was extracted according to Guillemaut et al. (1992), except for
RPC-5 column chromatography which was replaced by phenol-chloroform extraction.
2.3. PCR
The PCR assay of the NTS of the 5S rDNA was adopted from Kolchinsky et
al. (1991) and Cox et al. (1992).
2.4.
Polyacrylamide gel electrophoresis
The amplified fragments were separated under denaturing conditions on a
horizontal 10 % polyacrylamide gel (Mini CleanGel; Pharmacia Biotech, Freiburg,
Germany) with an amphoteric buffer system (DELECT forte buffer, pH 7.3; ETC-
Electrophorese Technik, Kirchentellinsfurt, Germany). The DNA fragments were
visualized by silver staining as described by Bassani et al. (1991).
3. Results
PCR amplification of NTS of avocado generated a variety of DNA fragments
as revealed by polyacrylamide gel electrophoresis with subsequent silver staining
(figure 1). The size of the PCR products varied from 200 bp to over 1000 bp.
Inferences from Southern hybridizations (Bufler and Ben-Ya'acov, 1992, and
unpublished results) suggest that fragments bigger than 800 bp are not
representing true NTSs. Thus, in the present study NTS sizes of avocado
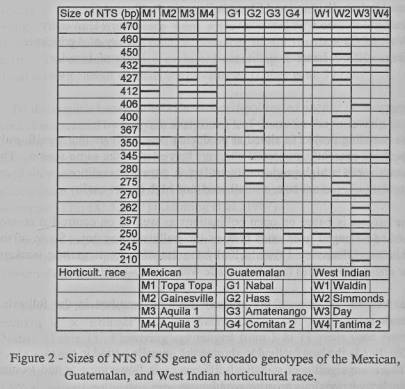
generated by PCR ranged from 210 bp to 470 bp (figures 1 and 2).
A comparison
of NTS sizes of 47 accessions of the subgenus Persea collected in Mexico
and Central America included cultivated avocados, wild relatives of avocado and
the semi-domesticated P. schiedeana. In this survey, NTS sizes common to
each horticultural race of avocado could be identified. The results of 12
representative avocado genotypes are summarized in figure 2. It shows, for
example, that Guatemalan and West Indian genotypes have a 470 bp and a 345 bp
NTS which is not found in Mexican genotypes. Some Guatemalan genotypes exhibit
NTSs of 245 bp and 250 bp in addition (or instead of, not shown) to the 345 bp
NTS. The NTSs at 406 bp and (in most cases) also at 412 bp are common to
Mexican race genotypes. The presence of the 432 bp NTS in genotypes of the Mexican or West Indian race
allows to distinguish between the West Indian and Guatemalan race. Thus, the
horticultural race of each genotype could be identified by the presence (or
absence) of certain NTS variants. Some cvs., namely 'Topa Topa', 'Gainesville',
'Hass', 'Waldin', 'Simmonds' and 'Day, contain NTS variants common to at least
two races and, therefore, are not considered as being 'pure' to a certain race
(figure 2; cf. Lahav and Gazit, 1994). Moreover, the cvs. 'Hass', 'Day' and
'Simmonds' exhibit NTS variants rarely found in other genotypes of the
cultivated avocado (figure 2).
4. Discussion
In a previous study,
Bufler and Ben-Ya'acov (1992) reported a 5S rDNA repeat size based on Southern
hybridization of 560 bp for Guatemalan and West Indian race avocado, and 540 bp
for Mexican race avocado. These results correspond reasonably well with the NTS
size of 470 bp for Guatemalan and West Indian race avocado and 460 bp for
Mexican race avocado if 80 bp are added for the transcribed region (the
remaining 40 bp represent binding sites for the 'Universal' primers, but become
part of the NTS after amplification. Thus, to estimate the correct size of a
NTS, 40 bp should be subtracted). Other repeat sizes are hardly detectable by
Southern hybridization.
Fumier et al. (1990) and Bufler and Ben-Ya'acov (1992) examined
restriction fragment length polymorphisms (RFLPs) of 18S-25S rDNA of avocado,
with the latter authors also reporting on the repeat size variation of 5S rDNA.
In these studies enough variation of rDNA could be observed to distinguish the
horticultural races of avocado. However, compared to the PCR-based assay of
NTSs the Southern hybridization-based assays are more laborious and time
consuming and less sensitive. Our results also demonstrate, that separation of
PCR products on a highly resolving polyacrylamide gel and highly sensitive
silver staining are prerequisites to fully exploit the variation of the NTSs of
avocado. Although it seems unlikely that the variation of NTSs of 5S rDNA gives
a good reflection of variation of the entire genome, it may provide a
diagnostic tool to identify the horticultural races of avocado cvs. and
germplasm collections. As a qualitative test, the NTS assay may also support
classification in the subgenus Persea and provide clues for phylogenetic
relationships.
Acknowledgements
We thank A. Ben-Ya'acov (Bet-Dagan, Israel) and A. Barrientos Priego
(Chapingo, Mexico) for providing avocado leaf samples. We appreciate the
financial support provided by the German-Israel Agricultural Research Agreement
(GIARA).
References
Bassam, B.J., Caetano-Anollós, G., and Gresshoff, P.M., 1991. Fast and
sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 196:
80-83.
Bergh, B., and Ellstrand, N., 1987. Taxonomy of the avocado. Calif.
Avocado Soc. Yrbk. 70: 135-145.
Bufler, G., and Ben-Ya'acov, A., 1992. A study of avocado germplasm
resources, 1988-1990. 111. Ribosomal DNA repeat unit polymorphism in avocado.
Proc. of Second World Avocado Congress. University of California, Riverside:
545- 550.
Cox, A. V., Bennett, M.D., and Dyer, T. A., 1992. Use of the polymerase
chain reaction to detect spacer size heterogeneity in plant 5S-rRNA gene
clusters and to locate such clusters in wheat (Triticum aestivwn L.). Theor.
Appl. Genet. 83: 684-690.
Furnier, G.R., Cummings, M.P., and Clegg, M.T., 1991. Evolution of the avocados
as revealed by DNA restriction fragment variation. J. Heredity 81: 183-188.
Guillemaut, P., and Mar6chal-Drouard, L., 1992. Isolation of plant DNA :
a fast, inexpensive, 'and reliable method. Plant. Mol. Biol. Rep. 10: 60-65.
Kolchinsky, A., Kolesnikova, M., and Ananiev, E., 1991. 'Portraying' of
plant genomes using polyrnerase chain reaction amplification of ribosomal 5S
genes, Genome 34: 1028-1031.
Lahav, E., and Gazit, S., 1994. World Listing of avocado cultivars
according to flowering type. Fruits 49: 299-313.
Sastri, D.C., Hilu, K., Appels, R., Lagudah, E.S., Playford, J., and
Baum, B.R., 1992. An overview of evolution in plant 5S DNA. Pl. Syst. Evol.
183: 169- 181.